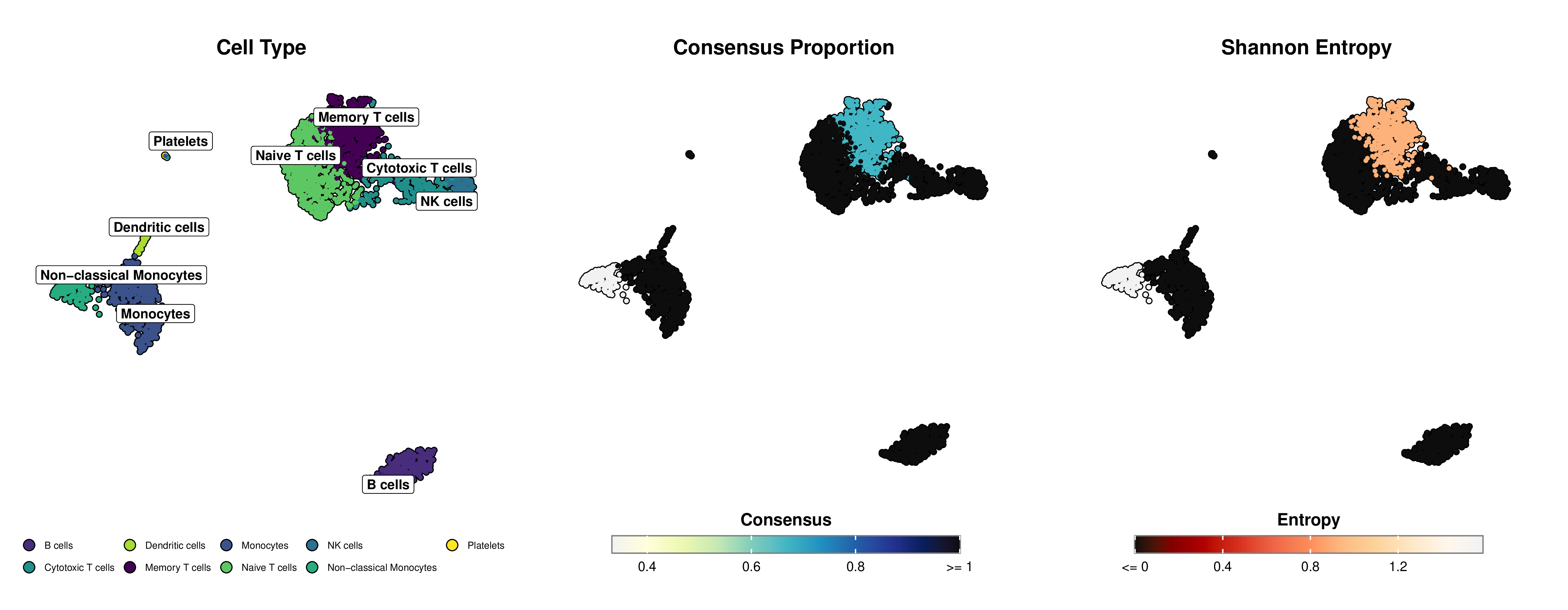
Multi-LLM Consensus Architecture for Cell Type Annotation in scRNA-seq Data
mLLMCelltype is an R package that leverages various large language models (LLMs) for automated cell type annotation in single-cell RNA sequencing data. The package implements a multi-LLM consensus architecture where multiple LLMs collaborate through structured deliberation that aims to improve annotation reliability by combining multiple model predictions.
Key Features
- Multi-LLM Consensus Mechanism: Combines predictions from multiple LLMs to reduce individual model biases
- Structured Deliberation Process: For controversial clusters, LLMs engage in collaborative discussion across multiple rounds, evaluating evidence and refining annotations together
- Uncertainty Quantification: Explicitly quantifies annotation uncertainty through consensus proportion and Shannon entropy
- No Reference Dataset Required: Does not rely on pre-existing reference datasets, can annotate various tissues and species
-
Support for Multiple LLM Providers:
- OpenAI (GPT-5.2, GPT-5, GPT-4.1, O3/O4 series)
- Anthropic (Claude 4.6 Opus, Claude 4.5 Sonnet/Haiku)
- Google (Gemini 3 Pro, Gemini 3 Flash, Gemini 2.5 series)
- X.AI (Grok-4, Grok-3 series)
- DeepSeek (DeepSeek Chat, DeepSeek Reasoner)
- Qwen (Qwen3 Max, Qwen Max)
- Zhipu (GLM-4.7, GLM-4 Plus)
- MiniMax (MiniMax M2.1, MiniMax M2)
- Stepfun (Step-3, Step-2 series)
- OpenRouter (access to Meta Llama, Mistral, Microsoft, Perplexity, Cohere, and more)
- Seurat Integration: Can directly use Seurat’s FindAllMarkers() output as input
Quick Start
# Install the package
devtools::install_github("cafferychen777/mLLMCelltype", subdir = "R")
# Load the package
library(mLLMCelltype)
# Set API keys
Sys.setenv(ANTHROPIC_API_KEY = "your-anthropic-api-key")
Sys.setenv(OPENAI_API_KEY = "your-openai-api-key")
Sys.setenv(GEMINI_API_KEY = "your-gemini-api-key")
# Use multiple models for annotation
models <- c(
"claude-3-7-sonnet-20250219",
"gpt-4o",
"gemini-2.5-pro"
)
# Run multi-model annotation
results <- list()
for (model in models) {
provider <- get_provider(model)
api_key <- switch(provider,
"anthropic" = Sys.getenv("ANTHROPIC_API_KEY"),
"openai" = Sys.getenv("OPENAI_API_KEY"),
"gemini" = Sys.getenv("GEMINI_API_KEY"))
results[[model]] <- annotate_cell_types(
input = pbmc_markers,
tissue_name = "human PBMC",
model = model,
api_key = api_key
)
}
# Create consensus using interactive consensus annotation
api_keys <- list(
anthropic = Sys.getenv("ANTHROPIC_API_KEY"),
openai = Sys.getenv("OPENAI_API_KEY"),
gemini = Sys.getenv("GEMINI_API_KEY")
)
consensus_results <- interactive_consensus_annotation(
input = pbmc_markers,
tissue_name = "human PBMC",
models = models, # Use the models defined above
api_keys = api_keys,
controversy_threshold = 0.7,
entropy_threshold = 1.0,
max_discussion_rounds = 3,
consensus_check_model = "claude-3-7-sonnet-20250219"
)Citation
If you use mLLMCelltype in your research, please cite our paper:
@article{Yang2025.04.10.647852,
author = {Chen Yang and Xianyang Zhang and Jun Chen},
title = {Large Language Model Consensus Substantially Improves the Cell Type Annotation Accuracy for scRNA-seq Data},
elocation-id = {2025.04.10.647852},
year = {2025},
doi = {10.1101/2025.04.10.647852},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2025/04/17/2025.04.10.647852},
journal = {bioRxiv}
}You can also cite this in plain text format:
Yang, C., Zhang, X., & Chen, J. (2025). Large Language Model Consensus Substantially Improves the Cell Type Annotation Accuracy for scRNA-seq Data. bioRxiv. https://doi.org/10.1101/2025.04.10.647852
Learn More
Please check our documentation to learn more about mLLMCelltype.